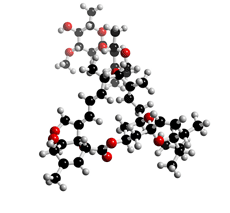
The molecular structure of Avermectin
Click on the image above to interact with the 3D model of the Avermectin structure
Avermectin
Discovery, producing organism and structure (1,2)
The discovery of avermectins was the result of a collaboration with Merck Sharp & Dohme Research Laboratories. Natural products were screened for anthelmintic activity using a direct in vivo test in mice infected with the nematode, Nematospiroides dubius. Avermectins were isolated from the culture broth of the actinomycete strain MA-4680T. Avermectins are a family of four closely-related major components, A1a, A2a, B1a and B2a, and four minor components, A1b, A2b, B1b and B2b, which are lower homologs of the corresponding major components. The first total synthesis of avermectin A1a was achieved by Danishefsky et al(3). Scince then, the total synthesis of avermectins has been reported by many groups.
Physical data ((Avermectin B1a)
White powder. C48H72O14; mol wt 873.08. Sol. in CHCl3, acetone, MeOH. Insol. in H2O.
Biological activity (1,4)
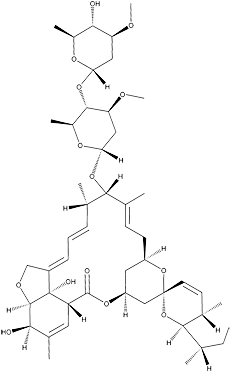
Avermectin
Avermectins have potent anthelmintic and insecticidal activities. In particular, the hydrogenated product of avermectin B1, 22,23-dihydroavermectin B1 (ivermectin), is used as an important anthelmintic in veterinary medicine as well as for the control of onchocerciasis, lymphatic filariais, strongyloidiasis and scabies in humans. The hydroxyl group at the C5 position and the disaccharide moiety are essential for the potency of avermectins.
New metabolites related to avermectin (5-7)
The following novel avermectin metabolites were obtained from blocked mutants of the producer.
Biosynthesis (8-18)
Several kinds of genes are involved in avermectin biosynthesis, aveA1, ave A2, ave A3, ave A4 (polyketide synthase), aveB (glycosylation), aveC (C-22,23 dehydration?), aveD (C-5 O-methylation), aveE (C-6, 8a furan ring formation), aveF (C-5 keto reduction), and aveR(regulation), were characterized and mapped on the chromosome.
Mode of action (19-23)
The target of avermectin is a glutamate-gated chloride channel.
Genome analysis (24, 25)
The entire genome of the avermectin-producing strain, Streptomyces avermectinius (avermitilis), has been mapped. It is the largest reported among the bacteria and represents the first complete genetic analysis of any industrially-important actinomycete.-----
Avermectin and Ivermectin are commercially available as anthelmintic agents for both
human and animals and used in agriculture.
References
- [155] R. W. Burg et al., Antimicrob. Agents Chemother., 15, 361–367 (1979)
- [821] Y. Takahashi et al., Int. J. Syst. Evol. Microbiol., 52, 2163-2168 (2002)
- S. J. Danishefsky et al., J. Am. Chem. Soc., 111, 2967–2980 (1989)
- K. S. Todd et al., Am. J. Vet. Res., 45, 976–977 (1984)
- [564] C.-H. Pang et al., J. Antibiot., 48, 59–66 (1995)
- [566] C.-H. Pang et al., J. Antibiot., 48, 92–94 (1995)
- [567] H. Ikeda et al., J. Antibiot., 48, 95–97 (1995)
- [381] H. Ikeda et al., J. Bacteriol., 169, 5615–5621 (1987)
- [388] H. Ikeda et al., Antimicrob., Agents Chemother., 32, 282–284 (1988)
- [462] S. Ōmura et al., J. Antibiot., 44, 560–563 (1991)
- [466] H. Ikeda and S. Ōmura, Kitasato Arch. Exp. Med., 63, 143–155 (1990)
- [476] H. Ikeda and S. Ōmura, Actinomycetol., 5, 86–99 (1991)
- [504] H. Ikeda et al., J. Bacteriol., 175, 2077–2082 (1993)
- [526] H. Ikeda and S. Ōmura, Actinomycetol., 7, 133–144 (1993)
- [575] H. Ikeda et al., J. Antibiot., 48, 532–534 (1995)
- [577] H. Ikeda and S. Ōmura, J. Antibiot., 48, 549–562 (1995)
- [685] H. Ikeda and S. Ōmura, Chem Rev., 97, 2591–2609 (1997)
- [693] H. Ikeda et al., Gene, 206,175-180 (1998)
- A. Pomes et al., Biochim. Biophys. Acta., 1339, 233–238 (1997)
- D. F. Cully et al., J. Biol. Chem., 271, 20187–20191 (1997)
- [722] H. Ikeda et al., Proc. Natl. Acad. Sci. USA, 96, 9509-9514 (1999)
- H. Ikeda et al., Actinomycetol., 13, 94-112 (1999)
- [790] H. Ikeda et al., J. Ind. Microbiol., 27, 170-176 (2001)
- [791] S. Ōmura et al., Proc. Natl. Acad. Sci., USA, 98, 12215-12220 (2001)
- [833] H. Ikeda et al., Nature Biotechnol., 21, 526-531 (2003)


